- 1Department of Arctic and Marine Biology, Faculty of Biosciences, Fisheries and Economics, UiT-The Arctic University of Norway, Tromsø, Norway
- 2Department of Microbiology, Institute for Biochemistry and Biology, University of Potsdam, Potsdam, Germany
Liverwort Blasia pusilla L. recruits soil nitrogen-fixing cyanobacteria of genus Nostoc as symbiotic partners. In this work we compared Nostoc community composition inside the plants and in the soil around them from two distant locations in Northern Norway. STRR fingerprinting and 16S rDNA phylogeny reconstruction showed a remarkable local diversity among isolates assigned to several Nostoc clades. An extensive web of negative allelopathic interactions was recorded at an agricultural site, but not at the undisturbed natural site. The cell extracts of the cyanobacteria did not show antimicrobial activities, but four isolates were shown to be cytotoxic to human cells. The secondary metabolite profiles of the isolates were mapped by MALDI-TOF MS, and the most prominent ions were further analyzed by Q-TOF for MS/MS aided identification. Symbiotic isolates produced a great variety of small peptide-like substances, most of which lack any record in the databases. Among identified compounds we found microcystin and nodularin variants toxic to eukaryotic cells. Microcystin producing chemotypes were dominating as symbiotic recruits but not in the free-living community. In addition, we were able to identify several novel aeruginosins and banyaside-like compounds, as well as nostocyclopeptides and nosperin.
Introduction
Multicellular nitrogen-fixing cyanobacteria of the genus Nostoc are a common component of terrestrial microbial communities in a wide range of habitats, including subpolar and hot arid zones. They are characterised by a versatile physiology and display one of the most complex life cycles among the bacteria. Nitrogen fixation in these microorganisms is confined to specialized cells, heterocysts, which appear in semi-regular pattern between chains of vegetative cells, where the photosynthesis takes place. In addition, the life cycle of Nostoc includes resting cells, akinetes, and motile multicellular filaments hormogonia, the dispersal units. Heterocystous cyanobacteria, primarily from the genus Nostoc are also engaged in a number of symbiotic interactions with plants and fungi (Meeks et al., 2002; Adams and Duggan, 2008). In such interactions, motile hormogonia serve as the infection units, whose differentiation is triggered by the host at initial stages of the infection process. The cyanobionts are housed either outside cells, in specialized cavities in the talli, as in liverworts and hornworts, or in roots as in cycads, or intracellular in symbiotic organs as in the angiosperms Gunnera sp. (Johansson and Bergman, 1992) or fungi of the genus Geosyphon (Kluge and Kape, 1992; Kluge, 1994). In these symbioses, both the hosts and the cyanobacterial partners are rather promiscuous, when the plant partners accept a wide range of Nostoc and a Nostoc strains isolated from one plant species is capable of infection phylogenetically distant hosts (Johansson and Bergman, 1994). The symbiotically competent Nostoc isolates were found along the entire Nostoc lineage of cyanobacteria (Papaefthimiou et al., 2008a) and do not show any patterns suggesting host specialization. The only exception is water-fern Azolla-Nostoc symbiosis, where the association is obligate and species specific for both and the partners are inseparable through the entire life cycle of the host (Papaefthimiou et al., 2008b; Zheng et al., 2009). The liverworts, hornworts and Gunnera produce the symbiotic structure before infection, and motile Nostoc filaments, hormogonia, enter these structures’ slime pores (Duckett et al., 1977; Rodgers and Stewart, 1977). Upon internalization, the heterocyst formation is usually up-regulated in symbiotic associations from about 4-10% in free-living cyanobacteria, to 25-60% when in symbiosis, although it seems likely that at least some of the extra heterocysts are non-functional (Meeks and Elhai, 2002). So far, no benefit was revealed for cyanobacteria to enter symbiotic interactions. Rather, formation of excess heterocysts diminishes propagation potential of eventually released cyanobionts.
Heterocystous cyanobacteria, including Nostoc are known as a rich source of biologically active secondary metabolites (Dembitsky and Rezanka, 2005; Rezanka and Dembitsky, 2006). Several compounds were used as drug leads, of which tumor inhibitors cryptophycins reached to the stage II clinical trials (Field et al., 2014). Analysis of the genome of N. punctiforme PCC73102 alone showed presence of 11 gene clusters possibly involved in non-ribosomal peptide, polyketide, and hybrid peptide-polyketide metabolites, most of which are still unknown (Liaimer et al., 2011). Further, over ten cryptic bacteriocin gene clusters with yet unexplored potential were predicted from the genomic sequence (Wang et al., 2011). Our recent findings show that at least some of these products, a cryptic polyketide compound and nostopeptolides are involved in the life cycle regulation and transition from motile stage to vegetative growth (Liaimer et al., 2011, 2015). Plant partners have a great impact on the production of secondary metabolites, by both down-regulating biosynthesis of metabolites observed in free-living state, and by inducing production of a great number unknown products (Liaimer et al., 2015). Therefore, genus Nostoc, with its complex life cycle and diverse interspecies interactions, represents a unique model for uncovering biological roles of secondary metabolites and discovery of novel compounds.
In their natural habitats, cyanobacteria will find themselves competing for resources with other organisms sharing similar environmental requirements, which include other cyanobacteria, microalgae and even higher plants. To date, only a very limited number of the cyanobacterial allelochemicals has been identified and mainly from freshwater cyanobacteria (Leflaive and Ten-Hage, 2007; Leão et al., 2009). These include cyanobacterin, a chlorinated γ-lactone produced by Scytonema hofmanni that inhibits other cyanobacteria and green microalgae (Mason et al., 1982); fisherellin A, an eynediyne-containing photosystem II inhibitor produced by Fisherella muscicola (Hagmann and Jüttner, 1996); the hapalindoles, small metabolites that have been isolated from Hapalosiphon (Moore et al., 1984; Thanh Doan et al., 2000) that show inhibitory effects against several microorganisms and the nostocyclamides, relatively small cyclic peptides produced by Nostoc sp. that inhibit cyanobacteria and microalgae (Todorova et al., 1995; Jüttner et al., 2001). The allelochemicals play important roles in phytoplankton succession, bloom formation and competition; several studies have investigated environmental factors that may modulate allelopathic events. The production of allelochemicals are controlled by the presence of competitors (Kearns and Hunter, 2000; Vardi et al., 2002; Jang et al., 2007), bacteria that degrade allelochemical compounds (Hulot and Huisman, 2004), light intensity, temperature, nutrient levels and pH (De Nobel et al., 1998; Ray and Bagchi, 2001; de Figueiredo et al., 2006).
In this study, we investigated symbiotic isolates obtained from Blasia pusilla L. naturally occurring in the various habitats in Troms region, Northern Norway. In these endophytic associations, the Nostoc colonies are housed in special cavities located on the ventral surface of the gametophyte (Figure 1). At the first steps we sorted the isolates by STRR-genome fingerprinting, followed by phylogeny reconstruction. The main goal of this work was, however, the chemical diversity in the Nostoc community inside the plant and in the soil around it, with an emphasis on small peptides based on whole cell and growth medium MALDI-TOF MS chemical typing. In addition, the fragmentation patterns of most prominent peaks were analyzed for the identification of peptides, when possible. Thus, we have aligned the genetic and chemical profiles among symbiotically competent Nostoc. We also tested the interstrain interactions by allelopathy assays in order to find possible explanations for differences between symbiotic and free-living communities. Having in mind the applied aspects of cyanobacterial secondary metabolites, we tested the isolates’ ability to inhibit different human cell lines, including two types of human cancer cells.

FIGURE 1. Blasia pusilla L. in its natural habitat. Symbiotic cavities, auricles, housing Nostoc are encircled. Cyanolichens Peltigera sp. shares the same environment with B. pusilla.
Materials and Methods
Isolation and Cultivation of Cyanobacteria
The plant material was collected on the territory of a plant school on Kvaløya island (69,64° N 18,73°E) near Tromsø and at an inland location near Skibotn field station (69,28°N 20,50°E), Northern Norway. The distance between the two sampling sites is approximately 81 km. Isolation of nitrogen-fixing cyanobacteria from soil and plants symbiotic organs was carried out as described earlier (West and Adams, 1997). The soil isolates were picked from the serial dilution plates. In order to suppress growth of algae and fungi the medium was supplemented with 20 mg/L cycloheximide for the first steps of isolation. All isolates were cultivated at room temperature in liquid or solidified BG110 medium (Stanier et al., 1971) under continuous illumination with light intensity of 60 μmol/sec∗m2.
Genotyping, Sequencing
The cyanobacterial samples were pipetted onto Whatman FTA classic cards (Sigma-Aldrich) and treated according to manufacturer’s instructions thereafter. STRR fingerprinting using 1, 2 mm FTA paper circles was performed according to Rasmussen and Svenning (1998). The first two letters in isolate label stand for location, KV-for Kvaløya, and SK for Skibotn, third letter J or S are random letters separating plant individuals, and F stands for free-living isolates from soil. The numbers stand for the order in which the isolate was picked from a given sample. Amplification of 16S rDNA and sequencing were done as described in Elvebakk et al. (2008). Sequences of 16S rDNA were deposited in the database of National Center for Biotechnology Information, NCBI, and assigned accession numbers EU022706 to EU022742 (Supplementary Table S1).
Alignment and Phylogenetic Analysis
Twenty-five 16S rRNA gene sequence of the genotypes defined in this study and 45 additional cyanobacterial 16S rRNA gene sequences available in the NCBI database (Supplementary Table S1) were used in the phylogeny reconstruction. The total data set of 16S sequences was aligned (Supplementary Figure S5) with the computer program Bioedit (Hall, 1999), producing sequence alignment which correspond to the Escherichia coli sequence region between positions 26 to 1522 (Neefs et al., 1990). The MEGA6 software program was used making phylogenetic analysis (Tamura et al., 2007). The tree was constructed based on 16S rRNA gene sequences by using the neighbor-joining method (NJ) algorithm with bootstrap values calculating from 1000 iterations. The Kimura two-parameter model was used to estimate evolutionary distance among the species (Kimura, 1980). Three non-heterocystous cyanobacterial 16S rRNA gene sequences (from Arthrospira platensis, Lyngbya sp. PCC 7419, Oscillatoria sp.) were used as outgroup.
The software program DOTUR (Schloss and Handelsman, 2005) was used to define operational taxonomic units (OTUs) at a threshold of 98% sequence similarity on the distance matrix obtained with DNAdist from the Phylip 3.66 package (Felsenstein, 1993).
Bioactivity Assays
Allelopathy assays were carried out as follows: a freshly plated lawn of indicator strain on solid BG110 agar (1,2 % agar) was overlaid by a thin layer of semisolid (0,7%) medium, inoculation loop sized agar blocks with tester strains were placed on the top agar. Petri dishes were kept under standard cultivation conditions until visible results, in most cases 2-3 weeks.
Cyanobacterial extracts for cell viability assays and antibacterial activity testing were obtained from cyanobacteria grown in 1 l medium under standard conditions. The cyanobacteria were collected by centrifugation at 6000 × g. The pellets were re-suspended in 50 ml methanol:water and sonicated with Sonifier250 (Branson); 3 cycles of 3 min each, 40% duty cycle, output control 3. The cell debris was removed by centrifugation. The extracts were dried under vacuum, washed out by 2 ml 50% methanol, and brought to dryness in 1,5 ml Eppendorf tubes. The resulted powder was dissolved to a concentration 50 mg/ml. The effect of cyanobacterial extracts on human cell viability was tested against two adherent cancer cell lines: human melanoma A2058 (American Type Culture Collection, ATCC CRL-11147) and human colon carcinoma HT29 (ATCC HTB-38). In addition, adherent, non-malignant lung fibroblasts MRC5 (ATCC CCL-171) were used as toxicity control. Initially a selection of extracts was tested against human melanoma A2058 with the crude extract concentration of 500 μg/mL. Four positively tested extracts were tried further against human melanoma A2058, human colon carcinoma HT29 and lung fibroblasts MRC5 with four concentrations ranging from 33 μg/mL to 250 μg/mL. Antibacterial activity was assayed against E. coli ATCC 25922, Staphylococcus aureus ATCC 25923, multi resistant S. aureus (MRSA) ATCC 33591 and Enterococcus faecalis ATCC 29212. The assays were performed at MabCent, University of Tromsø, Norway, following established procedures described in Hanssen et al. (2012) and Ingebrigtsen et al. (2016).
Mass Spectrometry
Lyophylized samples of cell pellets and supernatants were dissolved in water:acetonitril:ethanol (1:1:1) and analyzed by MALDI-TOF on a MALDI micro MXTM from Micromass Waters. A 10 mg/ml α-Cyano-4-hydroxycinnamic acid (CHCA) matrix in 49,5% acetonitril, 49,5% Etanol, and 10 μl 0,1% TFA was used. The instrument contained a nitrogen laser giving a 337 nm output. The ions were accelerated with a voltage of 20 kV. A delayed extraction of 500 ns and ion suppression up to 500 Da was used. The MS was used in the positive ion-detection and reflector mode. Data were collected using variable laser power, optimized for each sample. For tandem mass spectrometry samples containing peptides in water:acetonitril:ethanol (1:1:1) were added formic acid to 0.1%. The samples were analyzed in positive mode using a Q-TOF Ultima mass spectrometer (Micromass/Waters) with a nanospray source. Samples were injected with a flow of 0.5 μl/min. Mass spectra were acquired in continuum mode. The list of peptides of interest meant for the fragmentation analyses was manually defined in the equipment’s current settings. Collision energy for fragmentation was manually set for each peptide.
An outline over the workflow implemented in this study, covering steps from sample collection to bioactivity analyses is presented in Supplementary Figure S4.
Results
Isolation of Cyanobacteria, Genotyping, and Phylogeny Reconstruction
The two selected collection sites were chosen for their difference in history. Kvaløya site is an agricultural land, while Skibotn site is a natural habitat. In addition, the sites are separated from each other by natural hinders like fjords and mountains. Most of the cyanobiont colonies extracted from individual symbiotic organs, auricles, gave rise to free-living colonies on solid BG110 medium. All symbioses born colonies consisted of only one type of cyanobacteria, no mixed colonies were observed. The number of colonies to be taken for genotype sorting was limited to two hundred from each patch of B. pusilla, comprising in total approximately 800 isolates from four B. pusilla talli. All isolates were sorted into genotypes by STRR fingerprinting according to Rasmussen and Svenning (1998) (Supplementary Figure S1, Figure 2). In addition, the same amount of Nostocaceae colonies was collected from the soil to which talli were attached. Heterocystous cyanobacteria in soils were found at densities in a range from 0.5*104 to 6*106 CFU per cm3 soil. These values were not considered as accurate cell density estimates, since colonies in serial dilutions could rise from both single cells, as well as from multicellular filament or aggregates. The majority of the isolated cyanobacteria were successfully maintained in standard BG110 medium in liquid with the a few exceptions. Isolates from Kvaloya samples were assigned to 13 STRR-genotypes, and isolates from Skibotn were assigned to 12 genotypes, respectively (Supplementary Figure S1, Figure 2). Individual genotypes were designated with consecutive roman numbers. Sequencing of several random representatives from each genotype showed no variability in 16S sequences within an STRR-genotype, except for symbiotic genotype XI from Kvaloya and soil isolate XXI, which were first treated as the same genotype but were separated later due to differences in 16S rDNA sequences specific for each collection site. There were two immediate observations from the genotype sorting. Firstly, there were notable differences in the community composition between two sample sets from the same collection site. Very few symbiotic isolates were common for both plants from the same location, while the soil shared only the dominant genotypes but not the minor components (Figure 2). The second observation was that the symbiotic sets of genotypes were only partially overlapping with the soil sets of genotypes for all sites and patches. Indeed, most of the isolates found in symbioses were not found outside the plant. In three samples, Kvaloya1, Kvaloya2, and Skibotn2, genotypes which were the most abundant in planta, i.e., I, VI, and XXII, respectively, were not found in soil communities, whilst the dominating genotypes in soil, IV and V from Kvaloya and XX from Skibotn where not found as symbionts. However, the latter three were not assigned to Nostoc, but belonged to Calothrix, Trichormus, and Anabaena, respectively, according to 16S rDNA sequences and the phylogenetic tree (Supplementary Table S1, Figure 3). The remainder of the isolates was placed within Nostoc as shown in Figure 3, contained within two closely related OTUs. These two clades combined correspond to the “cluster I Nostoc” defined by Papaefthimiou et al. (2008a) and contain Nostoc sequences originated from various terrestrial ecosystems and a variety of symbiotic associations, ranging from lichens to Gunnera species. The only outlier in the phylogeny presented here was genotype XVIII, forming a distinct clade with an aquatic microcystin producing Nostoc sp. 152 (Sivonen et al., 1990).
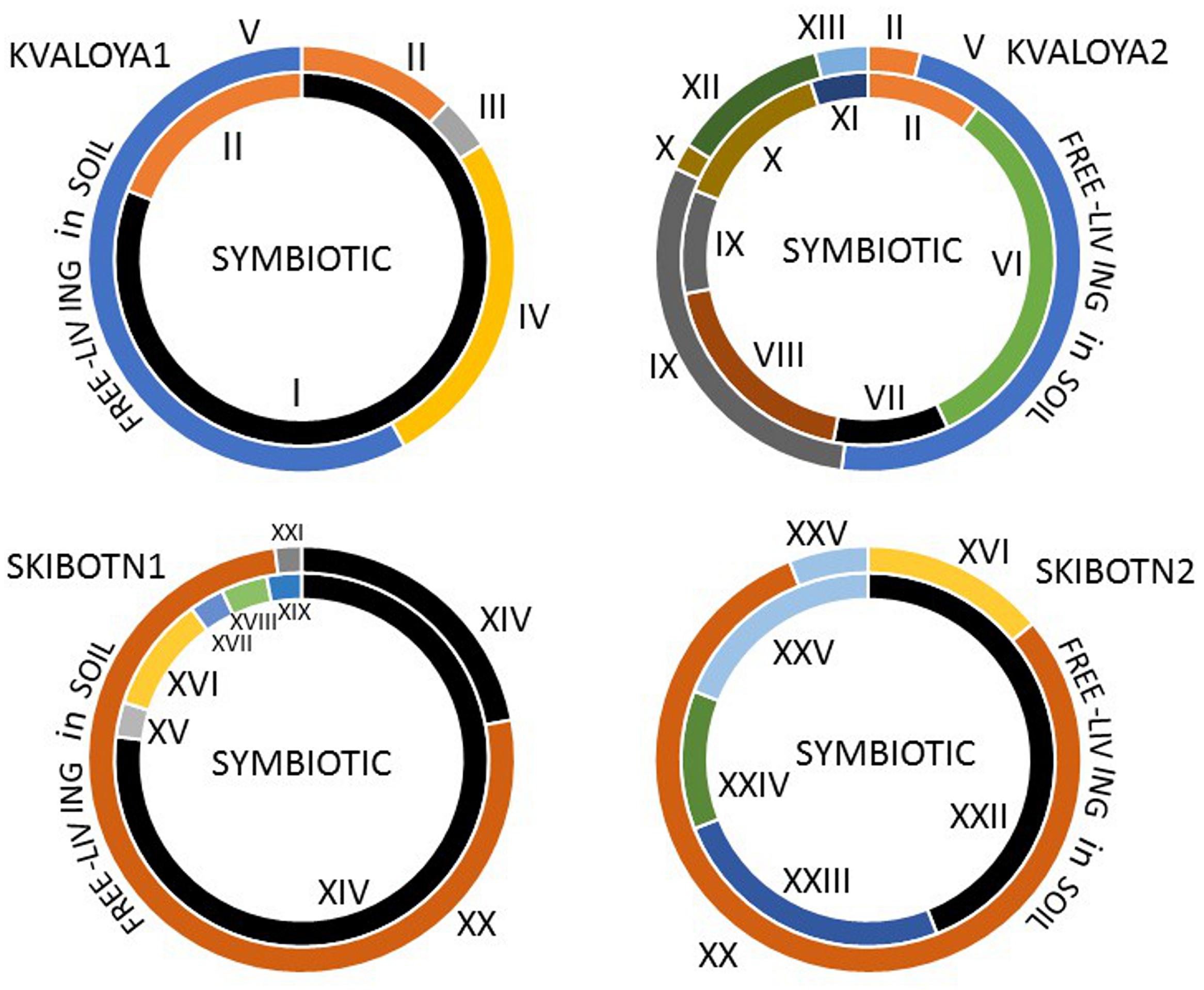
FIGURE 2. Distribution and abundance of Nostocacean STRR-genotypes in four sampled B. pusilla plants from two locations in Northern Norway, Kvaloya island, and Skibotn valley. The symbiotic genotypes are shown in inner circles, and the free-living genotypes found in soil around the plants are shown in outer circles. The genotypes producing microcystins are in black. The STRR- type fingerprints are shown in Supplementary Figure S1.
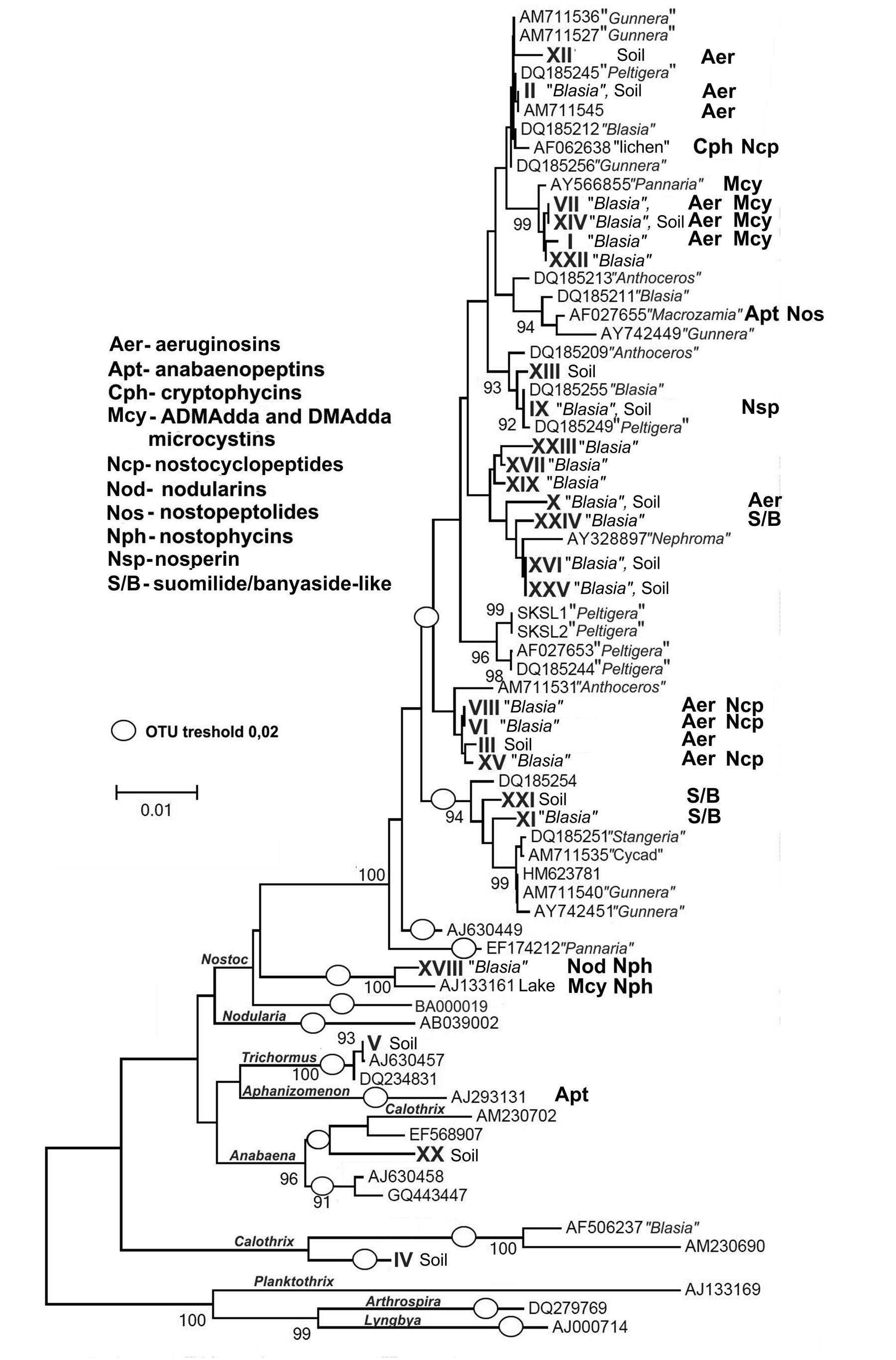
FIGURE 3. Phylogenetic tree showing the relationship among the 25 cyanobacterial STRR-genotypes found in this study (roman numbers) and 45 additional cyanobacterial 16S rRNA gene sequences. The tree was constructed based on 16S rRNA gene sequences by using the neighbor-joining method (NJ) algorithm with bootstrap values calculating from 1000 iterations. The Kimura two-parameter model was used to estimate evolutionary distance among the species (Kimura, 1980). Three non-heterocystous cyanobacterial 16S rRNA gene sequences (from Arthrospira platensis, Lyngbya sp. PCC 7419, Oscillatoria sp.) were used as outgroup. The scale bar corresponds to 0.01 nucleotide substitutions and (O) define operational taxonomic units (OTUs) at a threshold of 98% sequence similarity on the distance matrix. Distribution of secondary metabolites identified in this study or known from previous works are indicated next to the source strain.
MALDI-TOF MS Profiling and Metabolite Identification
Lyophilized cell pellets and filtered supernatants from all cultivable genotypes were subjected to MALDI-TOF MS analyses. In our preliminary studies we have noticed that lyophylization of the material prior to analyses aids to enhancement of peptide signals while the signals from chlorophyll and its derivatives stay low. We considered peaks with intensities over 5% relative to the highest observed. Our records include 254 individual peaks, representing 169 different compounds, in the range from 500 to 2000 Da. Several of the peaks were recognized as Na and K adducts commonly generated in ionization process in the MALDI-TOF MS analysis (Jemal et al., 1997; Mortier et al., 2004). The list of reproducibly occurring prominent peaks is presented in Table 1; the MALDI-TOF profiles are shown in Supplementary Figure S2. The most prominent ions were further subjected to fragmentation by Q-TOF MS. In this work we present fragmentation patterns (Supplementary Figure S3) of the compounds, which we were able to identify.
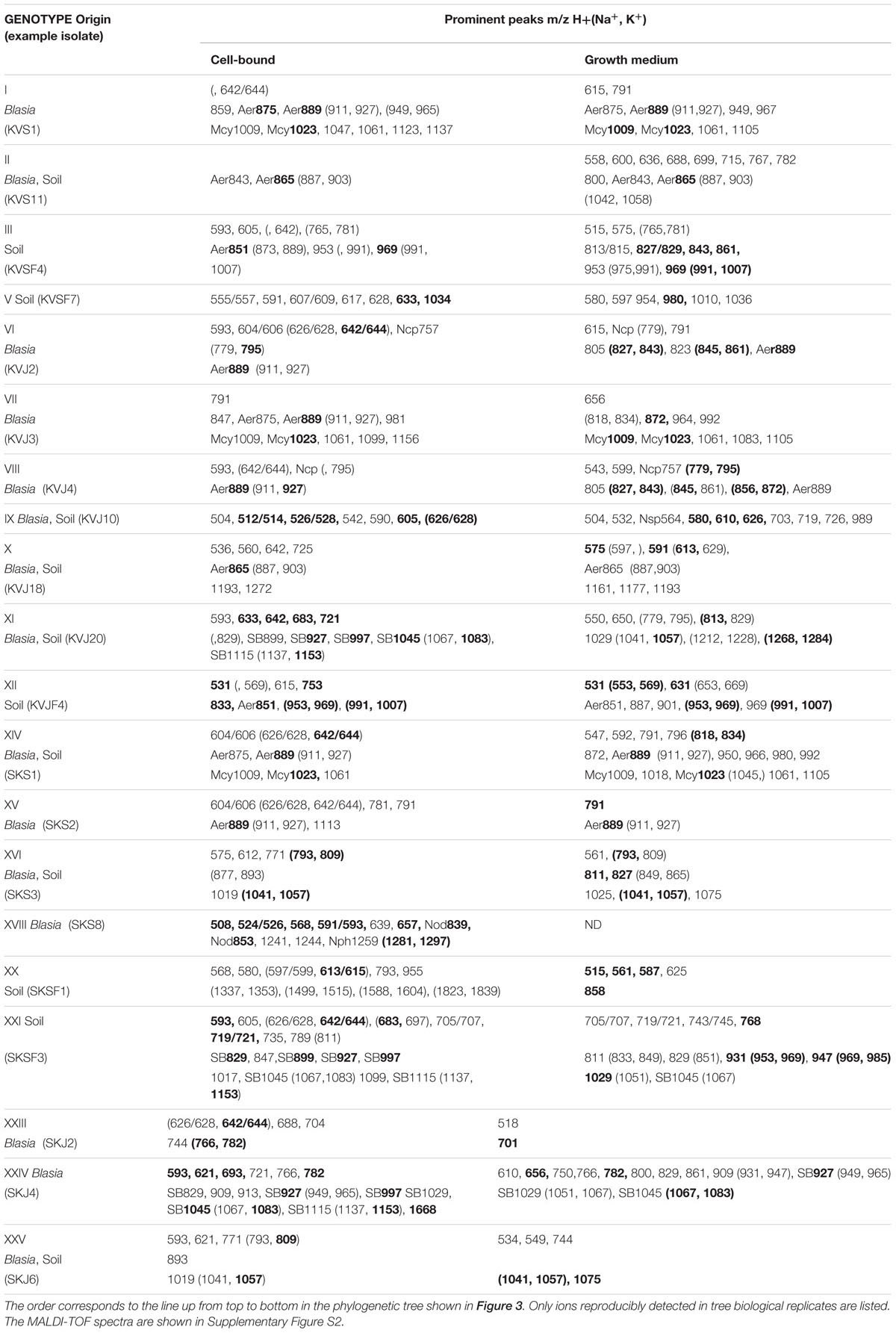
TABLE 1. Summary of the ions observed by MALDI-TOF screening of the cells and growth media of cyanobacterial isolates.
By conducting Q-TOF analyses followed by comparison with fragmentation patterns of known cyanobacterial metabolites, we were able to identify a number of metabolites (Table 2, Supplementary Figure S3).

TABLE 2. Summary of the compounds found in symbiotic Nostoc isolates assigned to known secondary metabolites produced by cyanobacteria.
Dominant ions m/z 1009 and 1023 produced by closely related I, VII, XIV, and XXII were identified as ADMAdda-microcystins found earlier in Nostoc from lichens and an aquatic Nostoc sp. 152. The fragmentation profiles matched the microcystin variants described in Oksanen et al., 2004, including diagnostic ions at m/z 155, 265, and 627 Da (Supplementary Figures S3G,H). Two nodularins with protonated ion masses 839 and 853 (Supplementary Figures S3I,J), were found in isolates of genotype XVIII (Nostoc sp. SKS8). These compounds were assigned due to combination of diagnostic ions m/z 209, 227, and 253 Da. The same strain produced a peptide with the mass m/z 1259, the fragmentation pattern of which was identical to that reported for nostophycin m/z 889 Da (Fujii et al., 2000) starting with the ion peak m/z 889 Da and lower. The fragmentation pattern suggests an additional side chain with three or four amino acids (Supplementary Figure S3K).
A peptide with m/z 865 has previously been reported from a number of terrestrial Nostoc strains (Hrouzek et al., 2011). The structure of the compound has been recently characterized as glucuronated aeruginosin 865 (Kapuscik et al., 2013). The fragmentation patterns of m/z 865 detected in this work in genotypes I, II, VI, VIII, and XIV and that of aeruginosin 865 were identical (Supplementary Figure S3B). Taking into account the overall similarity in MS/MS spectra of the remainder peptides showing loss of 176 or 162 Da, namely m/z 851 in gtIII, and m/z 875 and m/z 889 in several isolates, we may conclude that those compounds are also aeruginosins (Supplementary Figures S3A-D). The absence of fragment ions diagnostic of un-glycosylated aeruginosins in fragmentation patterns of glycosylated and glucuronated aeruginosins may be due to the influence of the side chains. A similar effect of side chains was described for ADMAdda-microcystins whose fragmentation patterns do not display diagnostic ions of more common and well-studied Adda-microcystins (Sivonen et al., 1990).
Isolates assigned to genotypes XI and XXI produced an array of substances ranging from m/z 829 Da to m/z 1045 Da sharing similar fragmentation patterns (Supplementary Figure S3L). The lower parts of fragmentation spectra were nearly identical to that of glycosidic peptide suomilide from Nodularia spumigena (Fujii et al., 2000). A prominent fragment ion of m/z 610 was present in all fragmentation spectra. An ion with this mass was also recorded in natural and synthetic banyaside, a peptide structurally similar to suomilide but produced in Nostoc (Schindler et al., 2010). The fragmentation patterns of the variants with masses over 1000 Da showed a neutral loss of 80 Da, corresponding to a sulfate group, also indicative of suomilide and banyasides (Fujii et al., 2000; Schindler et al., 2010).
Cell and medium extracts of several genotypes contained a peptide with protonated mass m/z 757 Da, but observed mostly as sodium and potassium adducts as m/z 779 Da and 795 Da. The fragmentation pattern displayed multiple losses corresponding to metPro, Gly, Gln, Ile, Ser, and Tyr or dipeptides composed of these amino acids (Supplementary Figure S3F), which proved to be the exact composition and molecular mass of nostocyclopeptide a1, found in lichen isolate Nostoc sp. ATCC53789 (Golakoti et al., 2001).
The last compound that could be identified was nosperin m/z 564 Da (Supplementary Figure S3E) found in supernatant of genotype IX (Nostoc sp. KVJ10), an unusual product of NRPS biosynthesis involving trans-acyltransferase originating from a lichen cyanobiont (Kampa et al., 2013).
The identified compounds observed in cell samples are also indicated on the phylogenetic tree (Figure 3). Examples of the chemical structures found in symbiotic Nostoc in this work are shown in Figure 4C.
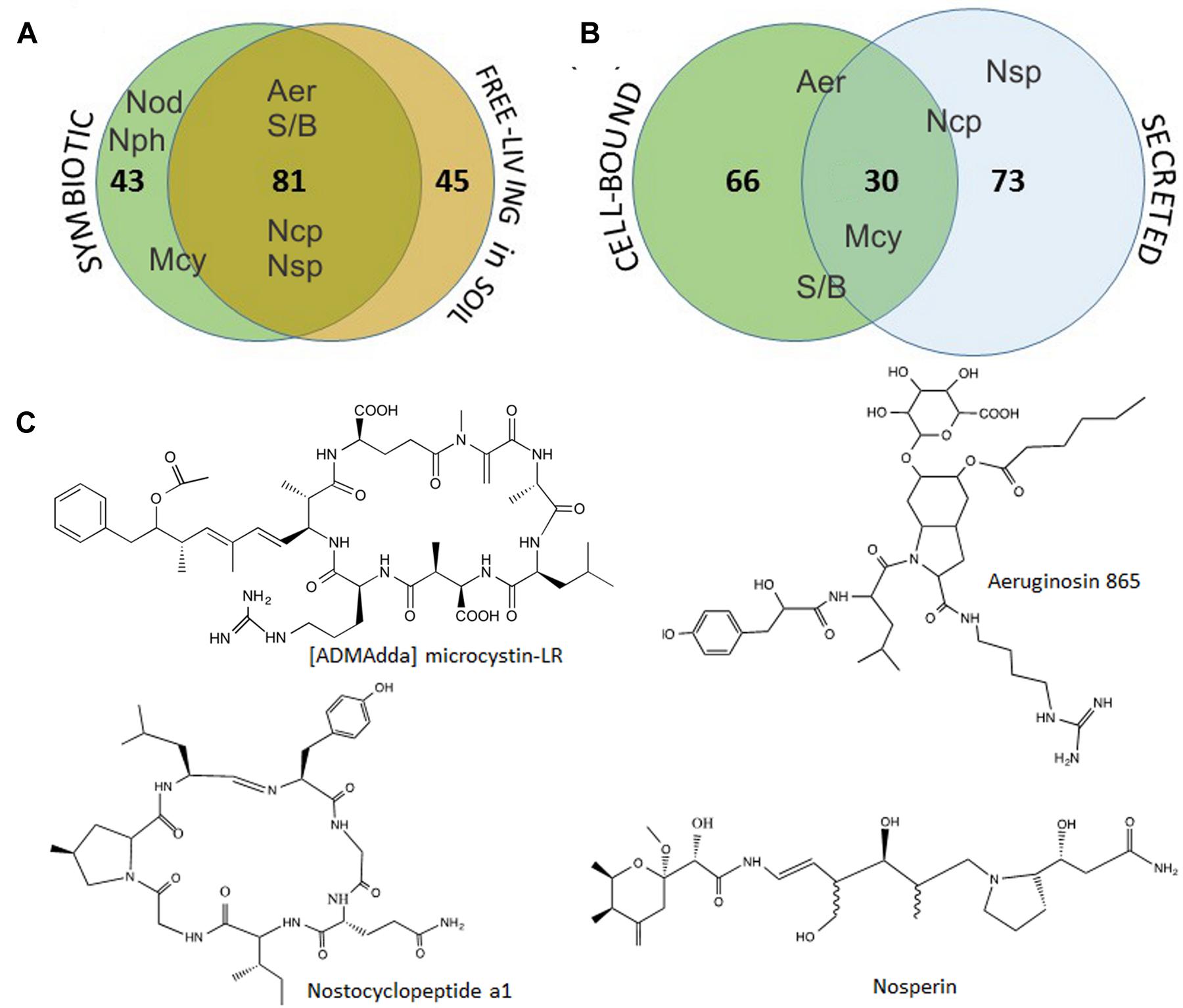
FIGURE 4. Cumulative distribution of the 169 metabolites detected in Nostoc isolates obtained from symbiotic association with B. pusilla or from soil at the sampling site (A), and the metabolites occurrence as cell-bound or secreted (B). Aer- aeruginosins, Mcy- [ADMAdda] microcystins, Ncp-nostocyclopeptide, Nod-nodularin, Nph-nostophycin, Nsp-nosperin, S/B-suomilide/banyaside-like compounds. The placement of the compound abbreviation shows it‘s tendency to be found as common for both sample sets/fraction or more specific to one. Selected examples of identified compounds, [ADMAdda] microcystin-LR, aeruginosin 865, nostocyclopeptide a1 and nosperin, are shown in (C).
Further, we have compared the compound lists found in symbiotic isolates and those found in soil. Nearly half of the metabolites (80 out of 169) were common for both groups of isolates. Moreover, all the major metabolites, including identified aeruginosins, suomilide/banyaside, nostocyclopeptide, and microcystins were found in both sets of samples (Figure 4A; Table 1). The differences in compound lists were largely due to peaks uniquely found only in particular strains. Producers of most major metabolites were equally represented in free-living and symbiotic sets of samples, except for microcystin and nodularin producers. Notably, microcystin producers were found in all four plant samples, but only in one soil sample (Skibotn1, Figure 2). Moreover, in the three plant samples Kvaloya1, Skibotn1 and 2, microcystin producing strains had the highest share. Nodularin producer was found only in symbiotic state in Skibotn1 sample (Figure 2).
When comparing metabolite lists from cell-extract with the secreted metabolome, we noted that the overlap between two lists is rather small. Out of 169 compounds only 30 where found in both fractions (Figure 4B). Analyzing MALDI-TOF profiles, we noted that microcystins, when produced, were found with equal intensities in both sets of samples. Aeruginosins and suomilide compounds were most pronounced in cell extracts, while the opposite was true for nostocyclopeptides (Figure 4B; Supplementary Figure S2). The sets of major cell-bound compounds were shared by several genotypes. While the peak lists of prominent extracellular metabolites were nearly always unique for each particular genotype, when the dually targeted compounds were subtracted (Table 1). This was especially pronounced in the mass range from 500 to 800 Da.
Bioactivities of Isolated Nostoc
We used two-layer agar bloc assay for detecting inhibitory activities among our isolates (Figure 5B). No pronounced inhibitory activity was found within the set of isolates from Skibotn. In contrast, cultivable Nostoc from Kvaloya site displayed a web of negative interactions between them (Figure 5A). Ten out of 12 cross-tested isolates showed inhibitory activity against at least one strain. Genotypes IX and VI showed the most powerful and broad range inhibition activity. Isolates with the most negative interactions directed against them were found only in planta. In order to rule out cyanophage infection as a cause of death, we have tested agar blocks from inhibition zones on new indicator lawn and did not observe any spreading (not shown). The cellular extract from the most active isolates, were tested on four clinical bacterial isolates (E. faecalis, E. coli, S. aureus, MRSA). The results of MIC test for all isolates were negative (not shown).
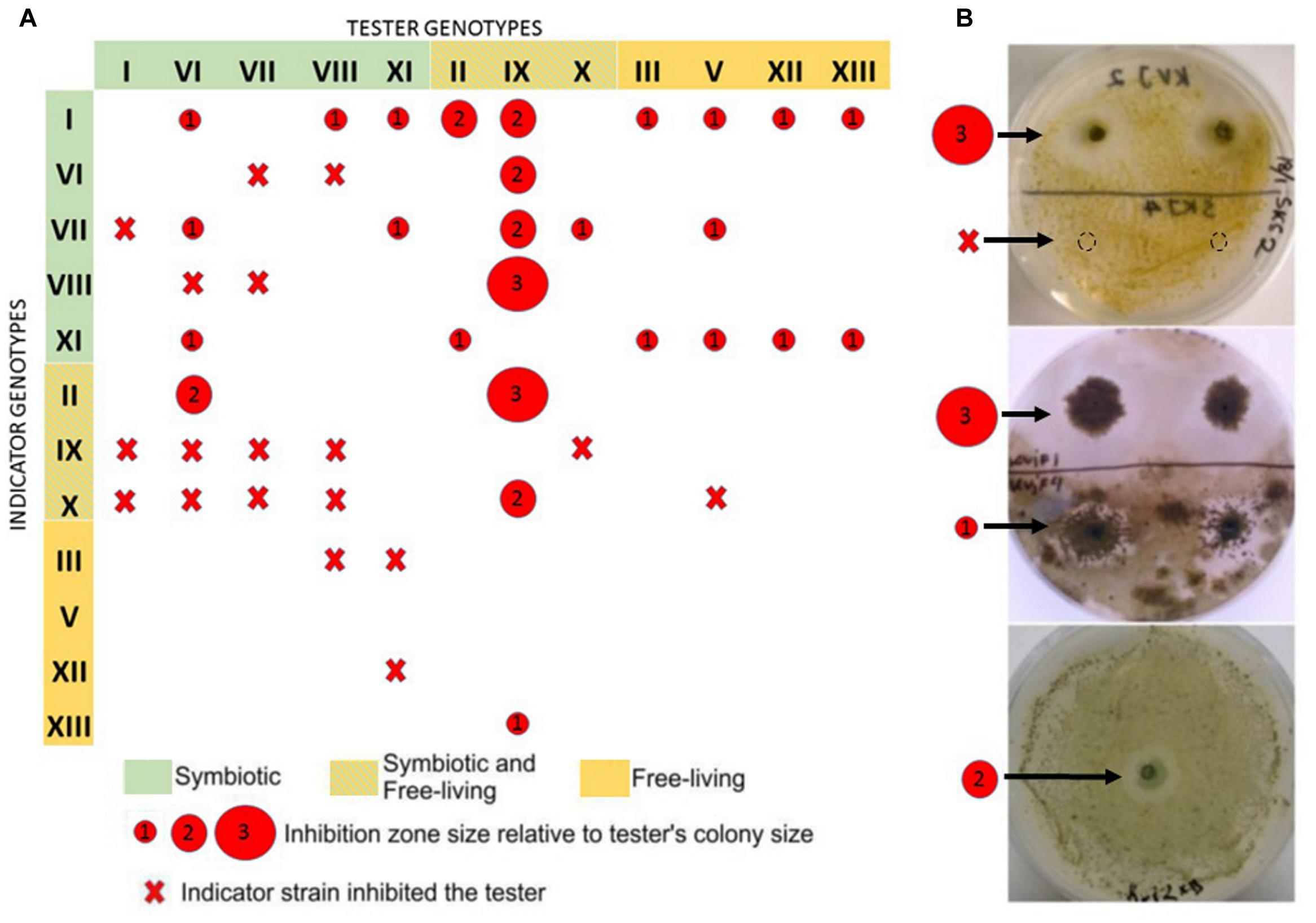
FIGURE 5. Presentation of allelopathic interactions between cyanobacterial isolates from the Kvaloya sampling site A. Summary of allelopathic interaction. (A) Summary table. The red circles represent the size of inhibition zone caused by the tester strains relative to its colonies radius, where 1 stands for inhibition zone up to 1- colony radius, 2- up to two radius, 3- from two radius sizes and larger. Red cross indicates that the indicator strain did not allow growth of the tester. (B) Examples of allelopathic assays conducted in this study.
Cytotoxicity tests were performed with total extracts from all isolates showing vigorous growth in culture. Initial screening was performed only with A2068 metastatic human melanoma cell line (Figure 6A). Extracts from four isolates, KVJ2 (genotypeVI), KVJ18 (X), KVJ20 (XI), and SKJ2 (XXIII) were notably inhibiting cell growth (Figure 6A). These isolates were further tested against MRC5 fibroblasts and HT29 human colon cancer cell lines. The latter was not effected by any of the extracts. MRC5 cells were inhibited in dosage dependent manner by KVJ20 and SKJ2 extracts (Figures 6B-D). Further characterization of cytotoxic properties of these strains is a subject of separate study.
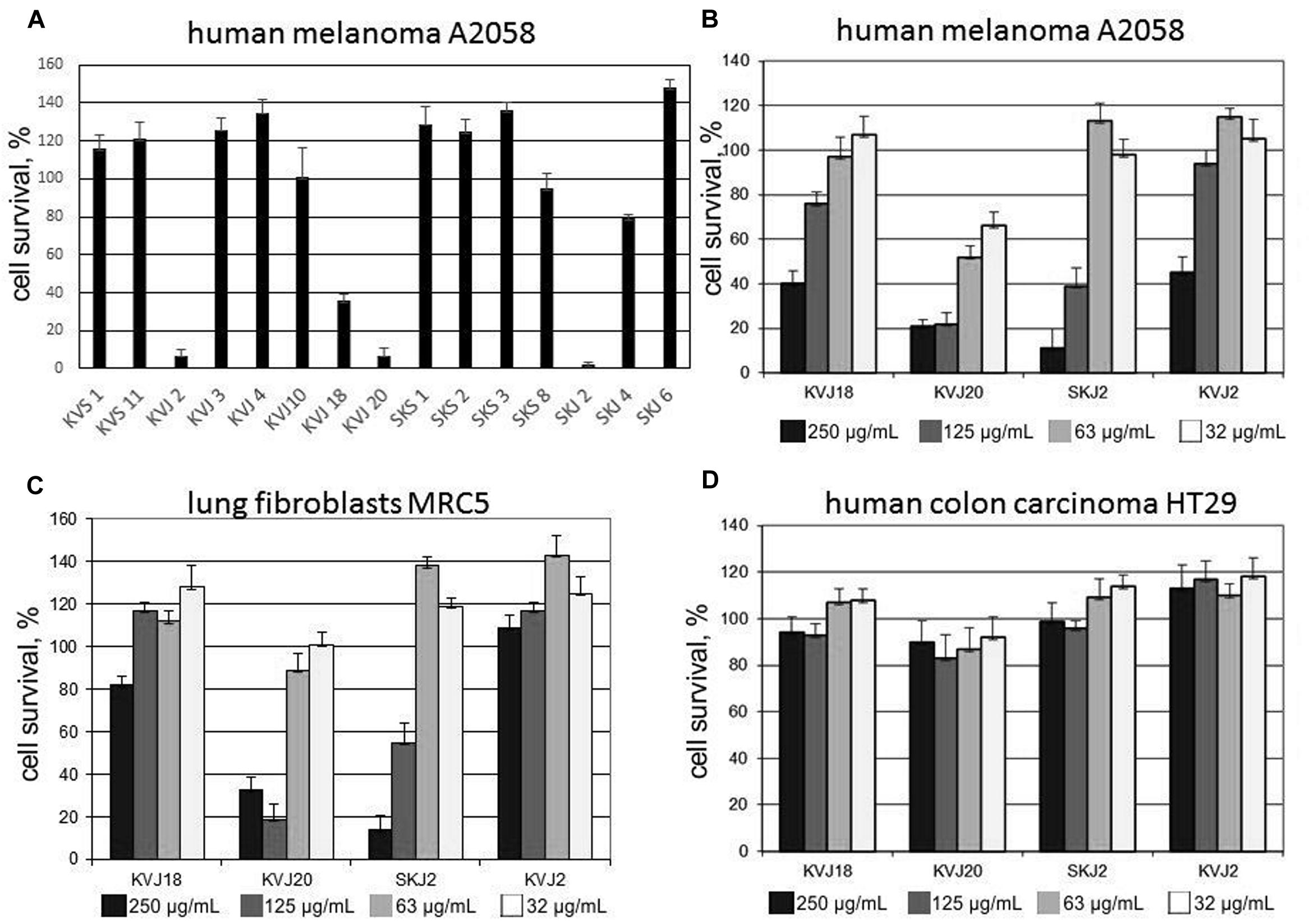
FIGURE 6. Cytotoxic activity of cell extract from a selection of Nostoc isolated in this study. (A) Cell survival test on human melanoma A2058 cell line applied 500 μg/mL crude extract from a selection of cyanobacterial isolates. (B-D) Cell survival tests performed with cell extracts of Nostoc sp. KVJ10, Nostoc sp. KVJ20, Nostoc sp SKJ2, and Nostoc sp. KVJ2 against human melanoma A2058, lung fibroblasts MRC5, and human colon carcinoma HT29 cell lines, respectively, at four different crude extract concentrations. Error bars-standard deviation of three technical replicates.
Discussion
Comments on the Genetic Diversity of Nostoc Recruited in Symbiosis
Here we report a difference in the cyanobiont diversity found even in two individual talli from the same location (Figure 2, Supplementary Figure S1). Our observations are in line with the previous report on cyanobacterial diversity in B. pusilla in England (West and Adams, 1997). The bryophyte host displayed promiscuity in recruitment of cyanobacterial partners, which were acquired locally. It is also likely that most of the cyanobacteria assigned to Nostoc would be able to act as a symbiont in this association. Several indications tell in support of general symbiotic competence in Nostoc. Most of the free-living isolates were able to infect B. pusilla under laboratory conditions (West and Adams, 1997). Free-living Nostoc isolates in our study are not phylogenetically distinct from symbiotic isolates, and it has been shown in several phylogeny reconstructions that symbiotic Nostoc are distributed through the entire Nostoc lineage (Enderlin and Meeks, 1983; Rasmussen and Svenning, 2001; Svenning et al., 2005; Papaefthimiou et al., 2008a). Epiphytic nitrogen-fixing cyanobacteria associated with bryophytes has been the topic of several recent publications and there is growing evidence that such associations are a widespread phenomenon (Bay et al., 2013; Sprent and Meeks, 2013). These new finding also tell in favor of symbiotic competence in genus Nostoc as rather general feature.
Nevertheless, our data also suggests that certain strains, microcystin producers in our case, are recruited more frequent than the others. Nilsson et al. (2005) have shown that in artificial association with rice roots, there was always one strain, which would outcompete others in the infection process. The feature was ascribed to faster hormogonia spreading, rather than ability to respond on plant elicited chemoattractant or inhibition of competitor strains. In natural population of Chilean Gunnera magelanica only one haplotype of Nostoc was found in an individual plant on the background of relatively high diversity of cyanobionts found in the population (Fernandez-Martinez et al., 2013). Obviously, there are yet unidentified selection processes taking place in each individual infection event. In our further discussion, we will argue that allelopathic interactions and the ability to produce certain secondary metabolites, at least in part, may be involved in shaping up symbiome in the host plant B. pusilla.
Allelopathic Interactions and Biological Activities of Cyanobacterial Extract
Allelopathic activities among cyanobacteria have been reported several times in the past. Therefore, we found it worthwhile to test whether the differences between free-living and symbiotic communities can be explained by such interactions. None of the extracts inhibited other bacteria, suggesting that the inhibitory action observed is rather specific against cyanobacteria. Thus, it is feasible to assume that allelopathic pressure from the free-living competitors can in part explain the differences between sets of genotypes found in symbiosis and outside. The absence of detectable allelopathic activity among Skibotn isolates can be explained by the fact that the site is a natural habitat with little disturbance from human activities, the Skibotn community thus can be considered as settled. In contrast, Kvaloya site is a part of a garden center where new soil with presumably new cyanobacteria is added every vegetation season, thus the observed interactions could reflect on-going succession in the soil community. Until now, there has not been pointed out a clear benefit for cyanobacteria in entering into symbioses with plants. Protection from negative allelopathic interactions may be one of such benefits; then an individual genotype outcompeted as free-living can be still present at a given location in a symbiotic state. Persistence of defined cyanobiont genotypes in B. pusilla through several years at the same location (Costa et al., 2001) tells in support of this hypothesis.
Cyanobacteria are considered as potentially valuable source of biologically active compounds for pharmaceutical applications (Singh et al., 2011; Tan, 2013). Hrouzek et al. (2011) previously reported the potential of symbiotic Nostoc as a source of cytotoxic compounds. In their screening of terrestrial Nostoc, 60% of strains of symbiotic origin possessed the property; however, the fraction of symbiotic isolates was very small in the total set. Therefore, we were tempted to conduct an initial cytotoxicity screening of Nostoc isolated in this study. Cytotoxicity tests were performed with total extracts from all cultivable isolates. Initial screening was performed only with A2068 metastatic human melanoma cell line. Extracts from four isolates, KVJ2 (genotypeVI), KVJ18 (X), KVJ20 (XI), and SKJ2 (XXIII) were notably inhibiting cell growth (Figure 6). These isolates were further tested against MRC5 fibroblasts and HT29 human colon cancer cell lines. The latter was not effected by any of the extracts, while both A2068 and MRC5 cells were inhibited in dosage dependent manner by KVJ20 and SKJ2 extracts (Figure 6). The results suggest a general cytotoxicity of KVJ20 and SKJ2. Further characterization of cytotoxic properties of these strains and their potential as sources of new drug lead compounds will be a subject of separate study.
Chemical Diversity of Symbiotic Nostoc
In this work we aimed to describe what bioactive potential symbiotic Nostoc have concerning their ability to produce small peptides, both associated with cellular material as well as secreted into the medium. Our results suggest that there is no obvious segregation, in regards of produced metabolites, between strains isolated from symbiotic association and from soil. Indeed, the cumulative overlap constituted nearly a half of the metabolites (Figure 4A). All observed major metabolites are included in the common set, including microcystins, aeruginosins nostocyclopetide, nosperin. For all samples combined, we noted that only 30 out of 169 recorded compounds are found in both cell-extracts and in the growth medium (Figure 4B). In fact, there are slightly more metabolites found as secreted. It can be speculated that indeed most of the small peptides are meant for secretion and are involved in extracellular functions such as communication, antibiotic activities and nutrient binding. Another immediate observation from peptide profiling of cells and media, is that many isolates have common sets of intracellular peptides (see Table 1). Such grouping became less apparent when the extracellular contents were compared. Most of the isolates show a unique extracellular peptide repertoire, especially in the range between 500 and 800 Da (Table 1). It is feasible to suggest that lower diversity and lower strain specificity in intracellular small peptides is related to their involvement in cellular processes, which are common and conserved among relatively close Nostoc isolates. Studies in quorum sensing and antibiotic production showed that secreted molecules, including small peptides, involved in communication and protection mechanisms, are often strain or species specific (Waters and Bassler, 2005; Fajardo and Martinez, 2008). A number of substances were found in both fractions. Even though there is a possibility of leakage from the cells, we would rather suggest that those substances are indeed truly dually targeted. This is supported by the fact that in the same samples, we have observed peaks that were detected exclusively in cells or vice versa only in supernatants (Table 1). Interestingly, the destination of the same compound may differ from isolate to isolate, when a compound in some strains is found solely as cell-bound while being dually targeted in others (Table 1).
In this study we did not aim to structurally elucidate individual peptides. However, we explored the possibility of assigning at least some of the compounds to previously described secondary metabolites (Table 2). We conducted extensive literature and database search in order to match observed masses to the existing records. The majority of the peaks could not be assigned to any candidate compound. In many cases when we would find a molecular weight match, the fragmentation patterns would not confirm our findings. We have also found that accumulated knowledge on cyanobacterial secondary metabolites is biased towards aquatic samples, which limited the possibility of precise identification in our study. There are still very few comprehensive studies of secondary metabolites in terrestrial (Hrouzek et al., 2011) or symbiotic cyanobacteria (Oksanen et al., 2004; Gehringer et al., 2010). Likewise, most of the compounds characterized in the literature, are obtained from the cell-extracts, creating a bias against secreted metabolites. This may be one of the reasons that almost none of the products found solely in the media was assigned to a known group of metabolites. This opens up an unexplored and prospective mine, since extracellular targeted biochemicals are most likely involved in interactions, and thus potentially possess activities desired, for example, in development of new antibiotics.
Nevertheless, the available reference data allowed us to identify a few previously reported compounds (Table 2, Supplementary Figure S3). Producers of cytotoxic microcystins and nodularins have been previously found in symbiotic associations in lichens and cycads (Oksanen et al., 2004; Gehringer et al., 2010). By identifying microcystins and nodularins in this work, we broaden the host range accepting potentially harmful cyanobionts. B. pusilla is a much better suitable model for studying the fate and regulation of cyanotoxins in association with eukaryotes. A further work with this interaction model may add more information on the biological role of microcystins and possible ways of manipulating their production.
There is an interesting group of compounds which include m/z 875 and 889 produced by genotypes I, VI, VII, XIV and XV, m/z 865 from II and X, and m/z 851 from III and XII. Fragmentation patterns of all these molecules share a common feature of a loss of either 176 or 162 Da, which is indicative of the presence of glucuronic acid or a hexose, respectively (Reinhold et al., 1995; Kapuscik et al., 2013). Glycosylation of non-ribosomal peptide and hybrid polyketide-peptide antibiotics in other bacteria is well known and was found to be important for the biological activity of such compounds (Walsh et al., 2001). Glycosylated aeruginosins described, until recently, exclusively in non-heterocystous cyanobacteria Planctothrix sp. were found in the same mass range as in our samples (Ishida et al., 2009). An antinflammatory peptide with m/z 865 has been previously reported from a number of terrestrial Nostoc strains (Hrouzek et al., 2011; Kapuscik et al., 2013). The structure of the compound was characterized as glucuronated aeruginosin-865 (Kapuscik et al., 2013). Our results show that production of aeruginosins is wide spread through Nostoc lineage. The very same is true for suomilide/banyaside like compounds, considered as related to aeruginosins (Pluotno and Carmeli, 2005).
Several Nostoc strains, found both in symbioses and in soil (genotypes VI, VIII, XI) produced nostocyclopeptide a1, which was found in cell material but was more pronounced in the media. Nostocyclopeptides are structurally similar to nostopeptolides, the gene cluster organization of Ncp and Nos operons share many common features, and there is a high degree of homology between key genes in the operons (Becker et al., 2004; Liu et al., 2014). This may suggest a similar biological function of the two compounds. In our recent work we showed that nostopeptolides are involved in regulation of the motile stage, hormogonia, acting as a suppressor of hormogonia formation (Liaimer et al., 2015), which may be also the function of nostocyclopeptides.
Phylogeny and Symbiotic Competence in the Context of Chemodiversity
In this study, we isolated over twenty genetically distinct symbiotic strains from genus Nostoc. Each of the isolate possessed a unique secondary metabolite profile. Each symbiotic organ, auricle, was infected by a single clone, which makes the plant partner to an ideal fishing devise for isolation of nearly clean cyanobacterial strains from soils.
During the last decade several groups attempted to find relationships between phylogenetic affiliation of cyanobacterial strains and their ability to produce certain secondary metabolites (Martin et al., 1993; Dawson, 1998; Ersmark et al., 2008). Microcystins, hepatotoxins that inhibit eukaryotic protein phosphatase activity, are produced by a variety of unicellular and filamentous cyanobacteria. Consequently, this group of compounds deserved a thorough attention also in the phylogeny studies. It has been suggested that microcystin gene cluster arose very early in cyanobacterial evolution (Rantala et al., 2004). The same authors hypothesized that the wide but mosaic distribution of mcy genes in most cyanobacterial lineages is most likely due to the loss of the function rather than repeated gain through the horizontal gene transfer. However, the possibility of the latter is not excluded and is debated (Mikalsen et al., 2003; Tooming-Klunderud et al., 2008). The long time prevailing hypothesis on the function of microcystins was protection from eukaryotic grazers and/or allelopathic action against eukaryotic competitors (Leflaive and Ten-Hage, 2007). However, the ancient origin of the gene cluster and predominantly intracellular localization of microcystins suggest that these functions are a secondary effect of these compounds. The latest evidence tells for possible involvement of microcystins in responses to oxidative stress (Zilliges et al., 2011). In our results, the microcystin producing genotypes, I, VII, XIV and XXII are found to be dominant in symbiosis, while not found outside the plant. These four isolates are phylogenetically related to the lichen cyanobiont (Figure 3) Our results also indicate that production of microcystins does not give a competitive advantage at these particular soil environments. Contrary to our expectations, the plant-host did not discriminate potentially toxic cyanobionts. For the majority of plant-microbe interactions, it has been reported that the host plants elicit an oxidiative burst during the early stages of infection processes (Mehdy, 1994; Wojtaszek, 1997). Even though this aspect of infection process in cyanobacterial-plant symbioses has not been elucidated, it is feasible to hypothesize that microcystin producers are better fit upon entry into the plant partner due to oxidative stress protective mechanisms aided by microcystins, and probably, structurally similar nodularins.
In this study we also showed a wide spread and greater diversity of aeruginosins, and related to them suomilide and banyaside-like compounds. These glycopeptides where found at nearly all branches on the Nostoc phylogeny, and are constituvely produced in culture, making this genus to a unique source of glycosylated NRPS products for bioactivity trials. Nostoc sp. KVJ10 (genotype IX) was found as a source of nosperin, an unusual NRPS product, belonging to pederin family. Secondary metabolites of this type has been described only from the microorganisms involved in symbiotic interaction (Kampa et al., 2013). Nostoc sp. KVJ10 and the cyanolichen isolate are to date the only source bacteria which are easily maintained in culture, and therefore represent a valuable model for investigation of biological role of nosperin, and pederin like compounds in general.
Due to the lack of reference data, we were not able to assign the majority of detected peptides to any known peptide classes. Cyanobacteria isolated from symbiotic association with plants and fungi were found to produce a wide spectrum of secondary metabolites. In addition to variety of microcystins from cyanolichens (Kaasalainen et al., 2012) and nodularins from a cycad (Gehringer et al., 2012) there were few other reports suggesting a broad repertoire of secondary metabolites and small peptides in symbiotic Nostoc. Nostopeptolides and anabaenopeptins were found in Nostoc punctiforme PCC73102 (Welker and von Dohren, 2006; Rouhiainen et al., 2010) originally isolated from Macrozamia sp., additionally this particular strain is rich in gene clusters coding for NRPS and hybrid NRPS-PKS metabolites (Ehrenreich et al., 2005) as well ribosomally synthesizes bacteriocin-like and microviridin products (Wang et al., 2011). Nostoc sp. ATCC53789 isolated from a cyanolichen has been extensively studied as a source of biologically active cryptophycins. A number of compounds belonging to all major cyanobacterial peptide classes have been described from free-living Nostoc (Rezanka and Dembitsky, 2006). From the mass spectra described in our study one may expect even greater range of unknown compounds with diverse biological activities to be discovered. Even though we noticed the dominance of microcystin producers in B. pusilla samples in this study, strains with very different metabolites are also accepted in symbiosis. At this point we may not say that there is a distinct cyanobiont selection pattern based on secondary metabolites identity. It implies that compounds potentially harmful for eukaryotic hosts are either contained within symbiotic organs, or their production is suppressed on regulatory or altered on biosynthetic level, or they are degraded by the plant. Symbiotic plant partners, such as B. pusilla and Gunnera sp. were shown to interfere with secondary metabolite production in N. punctifome (Liaimer et al., 2015). The fate of cyanobacterial small peptides in symbiotic interactions will definitely be the focal point of our further investigations. Based on our findings we suggest that the accessory features, secondary metabolites in particular, do not have great influence on symbiotic competence, rather, it is has an impact on a strains fate in the free-living community. It is likely that the cyanobiont discrimination is determined by the host’s ability to manipulate a particular strain, and the key to this manipulation lies in the core genome of the Nostoc. In either scenario, the mechanisms of such interactions are intriguing and may have application beyond the field of plant-cyanobacterial symbioses.
Conclusion
In this work we demonstrated a high genetic and chemodiversity of symbiotic nitrogen-fixing cyanobacteria in liverwort B. pusilla L. from two site in Northern Norway. We showed also that the host plant does not discriminate cyanotoxin producers, contrary, those were the dominant group in symbioses. The toxin production did not provide competitive advantage in free-living community. Cyanobacteria with strong allelopathic abilities can be a valuable source for compounds with cyanostatic activities. Additionally we showed that several isolates contain yet unidentified cytotoxic substances which can be used in anticancer drug development. Most of the peptide peaks observed in our study represent compounds with no records in the literature, thus making terrestrial cyanobacteria to perspective poorly explored source of novel natural products. For example, we showed a greater diversity and wide distribution of glycosylated NRPS products in this phylum. This study opens up for several intriguing question in regards to the fate and role of small peptides in plant symbioses.
Author Contributions
AL: Design of the concept and experimental approach, sample collection, isolation, and cultivation of strains, analyses of STRR fingerprints, sequencing, allelopathy trials, MALDI-TOF analyses, interpretation of MS/MS data, figure preparation, manuscript writing, and guiding technical personnel. JJ: Design of the concept and experimental approach, phylogeny reconstruction and interpretation, manuscript writing, figure preparation, and guiding technical personnel. ED: Design of the concept and experimental approach, interpretation of MS/MS data, literature search, figure preparation, and manuscript writing.
Funding
UiT provided basic funding.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Many thanks to Janne Stangeland and Eli Helene Robertsen AMB, BFE for technical assistance. Arve Elvebakk from Tromsø Botanical Garden, UiT, suggested the sampling sites for B. pusilla L. Toril Grønset and Jack Bruun at the Tromsø University Proteomics Platform (TUPP) are thanked for consulting on MALDI-TOF and Q-TOF analyses. MabCent at University of Tromsø is acknowledged for conducting antibacterial assays and cell viability tests.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01693
References
Adams, D. G., and Duggan, P. S. (2008). Cyanobacteria–bryophyte symbioses. J. Exp. Bot. 59, 1047–1058. doi: 10.1093/jxb/ern005
Bay, G., Nahar, N., Oubre, M., Whitehouse, M. J., Wardle, D. A., Zackrisson, O., et al. (2013). Boreal feather mosses secrete chemical signals to gain nitrogen. New Phytol. 200, 54–60. doi: 10.1111/nph.12403
Becker, J. E., Moore, R. E., and Moore, B. S. (2004). Cloning, sequencing, and biochemical characterisation of the nostocyclopeptide biosynthetic gene cluster: molecular basis for imine macrocyclization. Gene 325, 35–42. doi: 10.1016/j.gene.2003.09.034
Costa, J. L., Paulsrud, P., Rikkinen, J., and Lindblad, P. (2001). Genetic diversity of Nostoc symbionts endophytically associated with two bryophyte species. Appl. Environ. Microbiol. 67, 4393–4396. doi: 10.1128/AEM.67.9.4393-4396.2001
Dawson, R. M. (1998). The toxicology of microcystins. Toxicon 36, 953–962. doi: 10.1016/S0041-0101(97)00102-5
de Figueiredo, D., Reboleira, A., Antunes, S., Abrantes, N., Azeiteiro, U., Gonçalves, F., et al. (2006). The effect of environmental parameters and cyanobacterial blooms on phytoplankton dynamics of a Portuguese temperate Lake. Hydrobiologia 568, 145–157. doi: 10.1007/s10750-006-0196-y
De Nobel, W. T., Matthijs, H. C. P., Von Elert, E., and Mur, L. R. (1998). Comparison of the light-limited growth of the nitrogen-fixing cyanobacteria Anabaena and Aphanizomenon. New Phytol. 138, 579–587. doi: 10.1046/j.1469-8137.1998.00155.x
Dembitsky, V. M., and Rezanka, T. (2005). Metabolites produced by nitrogen-fixing Nostoc species. Folia Microbiol. 50, 3663–3691. doi: 10.1007/BF02931419
Duckett, J. G., Prasad, A. K. S. K., Davies, D. A., and Walker, S. (1977). A cytological analysis of the Nostoc-bryophyte relationship. New Phytol. 79, 349–362. doi: 10.1111/j.1469-8137.1977.tb02215.x
Ehrenreich, I. M., Waterbury, J. B., and Webb, E. A. (2005). Distribution and diversity of natural product genes in marine and freshwater cyanobacterial cultures and genomes. Appl. Environ. Microbiol. 71, 7401–7413. doi: 10.1128/AEM.71.11.7401-7413.2005
Elvebakk, A., Papaefthimiou, D., Robertsen, E. H., and Liaimer, A. (2008). Phylogenetic patterns among Nostoc cyanobionts within bi- and triparttite lichens of the genus Pannaria. J. Phycol. 44, 1049–1059. doi: 10.1111/j.1529-8817.2008.00556.x
Enderlin, C., and Meeks, J. (1983). Pure culture and reconstitution of the Anthoceros-Nostoc symbiotic association. Planta 158, 157–165. doi: 10.1007/BF00397709
Ersmark, K., Del Valle, J. R., and Hanessian, S. (2008). Chemistry and biology of the aeruginosin family of serine protease inhibitors. Angew. Chem. Int. Ed. Engl. 47, 1202–1223. doi: 10.1002/anie.200890018
Fajardo, A., and Martinez, J. L. (2008). Antibiotics as signals that trigger specific bacterial responses. Curr. Opin. Microbiol. 11, 161–167. doi: 10.1016/j.mib.2008.02.006
Felsenstein, J. (1993). Phylogeny Inference Package (PHYLIP). Version 3.5. Seattle, WA: Department of Genetics, University of Washington.
Fernandez-Martinez, M. A., de los Rios, A., Sancho, L. G., and Perez-Ortega, S. (2013). Diversity of endosymbiotic Nostoc in Gunnera magellanica (L) from Tierra del Fuego, Chile. Microbial Ecol. 66, 335–350. doi: 10.1007/s00248-013-0244-x
Field, J. J., Kankkanthara, A., and Miller, J. H. (2014). Microtubule-targeting agents are clinically succesfull due to both mitotic and interphase impairment of microtubule function. Bioorg. Medicin. Chem. 22, 5050–5059. doi: 10.1016/j.bmc.2014.02.035
Fujii, K., Mayumi, T., Noguchi, K., Kashiwagi, T., Akashi, S., Sivonen, K., et al. (2000). Mass spectrometric studies of peptides from cyanobacteria under FAB MS/MS conditions. J. Mass. Spectrom. Soc. Jpn. 48, 56–64. doi: 10.5702/massspec.48.56
Gehringer, M. M., Adler, L., Roberts, A. A., Moffitt, M. C., Mihali, T. K., Mills, T. J. T., et al. (2012). Nodularin, a cyanobacterial toxin, is synthesized in planta by symbiotic Nostoc sp. ISME J. 6, 1834–1847. doi: 10.1038/ismej.2012.25
Gehringer, M. M., Pengelly, J. J., Cuddy, W. S., Fieker, C., Forster, P. I., and Neilan, B. A. (2010). Host selection of symbiotic cyanobacteria in 31 species of the Australian cycad genus: Macrozamia (Zamiaceae). Mol. Plant. Microbe. Interact. 23, 811–822. doi: 10.1094/MPMI-23-6-0811
Golakoti, T., Yoshida, W. Y., Chaganty, S., and Moore, R. E. (2001). Isolation and structure determination of Nostocyclopeptides A1 and A2 from the terrestrial cyanobacterium Nostoc sp. ATCC53789. J. Nat. Prod. 64, 54–59. doi: 10.1021/np000316k
Hagmann, L., and Jüttner, F. (1996). Fischerellin A, a novel photosystem-II-inhibiting allelochemical of the cyanobacterium Fischerella muscicola with antifungal and herbicidal activity. Tetrahedron Lett. 37, 6539–6542. doi: 10.1016/0040-4039(96)01445-1
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Series 41, 95–98.
Hanssen, K. Ø, Andersen, J. H., Stiberg, T., Engh, R. A., Svenson, J., Geneviere, A. M., et al. (2012). Antitumoral and nechanistic studies of ianthelline isolated from the Arctic sponge Striphnus fortis. Anticancer. Res. 32, 4287–4297.
Hrouzek, P., Tomek, P., Lukesova, A., Urban, J., Voloshko, L., Pushparaj, B., et al. (2011). Cytotoxicity and secondary metabolites production in terrestrial Nostoc strains, originating from different climatic/geographic regions and habitats: is their cytotoxicity environmentally dependent? Environ. Toxicol. 26, 345–358. doi: 10.1002/tox.20561
Hulot, F. D., and Huisman, J. (2004). Allelopathic interactions between phytoplankton species: the roles of heterotrophic bacteria and mixing intensity. Limnol. Oceanogr. 49, 1424–1434. doi: 10.4319/lo.2004.49.4_part_2.1424
Ingebrigtsen, R. A., Hansen, E., Andersen, J. H., and Eilertsen, H. C. (2016). Light and temperature effects on bioactivity in diatoms. J. Appl. Phycol. 28, 939–950. doi: 10.1007/s10811-015-0631-4
Ishida, K., Welker, M., Christiansen, G., Cadel-Six, S., Bouchier, C., Dittmann, E., et al. (2009). Plasticity and evolution of aeruginosin biosynthesis in cyanobacteria. Appl. Environ. Microbiol. 75, 2017–2026. doi: 10.1128/AEM.02258-08
Jang, M.-H., Ha, K., and Takamura, N. (2007). Reciprocal allelopathic responses between toxic cyanobacteria (Microcystis aeruginosa) and duckweed (Lemna japonica). Toxicon 49, 727–733. doi: 10.1016/j.toxicon.2006.11.017
Jemal, M., Almond, R. B., and Teitz, D. S. (1997). Quantitative bioanalysis utilizing high-performance liquid chromatography/electrospray mass spectrometry via selected-ion monitoring of the sodium ion adduct [M+Na]+. Rapid Commun. Mass Spectrom. 11, 1083–1088. doi: 10.1002/(SICI)1097-0231(19970630)11:10<1083::AID-RCM937>3.0.CO;2-F
Johansson, C., and Bergman, B. (1992). Early events during the establishment of the Gunnera/Nostoc symbiosis. Planta 188, 403–413. doi: 10.1007/BF00192808
Johansson, C., and Bergman, B. (1994). Reconstitution of the symbiosis of Gunnera manicata Linden: cyanobacterial specificity. New Phytol. 126, 643–652. doi: 10.1111/j.1469-8137.1994.tb02960.x
Jüttner, F., Todorova, A. K., Walch, N., and von Philipsborn, W. (2001). Nostocyclamide M: a cyanobacterial cyclic peptide with allelopathic activity from Nostoc 31. Phytochemistry 57, 613–619. doi: 10.1016/S0031-9422(00)00470-2
Kaasalainen, U., Fewer, D. P., Jokela, J., Wahlsten, M., Sivonen, K., and Rikkinen, J. (2012). Cyanobacteria produce a high variety of hepatotoxic peptides in lichen symbiosis. Proc. Natl. Acad Sci. U.S.A. 109, 5886–5891. doi: 10.1073/pnas.1200279109
Kampa, A., Gagunashvili, A. N., Gulder, T. A., Morinaka, B. I., Daolio, C., Godejohann, M., et al. (2013). Metagenomic natural product discovery in lichen provides evodence for a family of biosynthetic pathways in diverse symbioses. Proc. Natl. Acad Sci. U.S.A. 110, 3129–3137. doi: 10.1073/pnas.1305867110
Kapuscik, A., Hrouzek, P., Kuzma, M., Bartova, S., Novak, P., Jokela, J., et al. (2013). Novel aeruginosin-865 from Nostoc sp. as a potent anti.inflammatory agent. Chembiochem. 14, 2329–2337. doi: 10.1002/cbic.201300246
Kearns, K. D., and Hunter, M. D. (2000). Green algal extracellular products regulate antialgal toxin production in a cyanobacterium. Environ. Microbiol. 2, 291–297. doi: 10.1046/j.1462-2920.2000.00104.x
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide-sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kluge, M. (1994). Geosiphon pyriforme (Kützing) von Wettstein, a promising system for studying endocyanoses. Prog. Bot. 55, 130–141.
Kluge, M., and Kape, R. (1992). Geosiphon pyriforme, an endosymbiotic consortium of a fungus and a cyanobacterium (Nostoc), fixes nitrogen. Bot. Acta 105, 343–344. doi: 10.1111/j.1438-8677.1992.tb00309.x
Leão, P. N., Vasconcelos, M. T. S. D., and Vasconcelos, V. M. (2009). Allelopathy in freshwater cyanobacteria. Crit. Rev. Microbiol. 35, 271–282. doi: 10.3109/10408410902823705
Leflaive, J., and Ten-Hage, L. (2007). Algal and cyanobacterial secondary metabolites in freshwaters: a comparison of allelopathic compounds and toxins. Freshwater Biol. 52, 199–214. doi: 10.1111/j.1365-2427.2006.01689.x
Liaimer, A., Helfrich, E. J. N., Hinrichs, K., Guljamow, A., Ishida, K., Hertweck, C., et al. (2015). Nostopeptolide plays a governing role during cellular differentiation of the symbiotic cyanobacterium Nostoc punctiforme. Proc. Natl. Acad Sci. U.S.A. 112, 1862–1867. doi: 10.1073/pnas.1419543112
Liaimer, A., Jenke-Kodama, H., Ishida, K., Hinrichs, K., Stangeland, J., Hertweck, C., et al. (2011). A polyketide interferes with cellular differentiation in the symbiotic cyanobacterium Nostoc punctiforme. Environ. Microbiol. Rep. 3, 550–558. doi: 10.1111/j.1758-2229.2011.00258.x
Liu, L., Jokela, J., Herfindal, L., Wahlsten, M., Sinkkonen, J., Permi, P., et al. (2014). Methylproline guided natural product discovery: co-occurrence of 4-Hydroxy- and 4-methylprolines in nostoweipeptins and nostopeptolides. ASC Chem. Biol. 9, 2646–2655.
Martin, C., Oberer, L., Ino, T., Konig, W. A., Busch, M., and Weckesser, J. (1993). Cyanopeptolins, new depsipeptides from the cyanobacterium Microcystis sp. PCC 7806. J. Antibiot. (Tokyo) 46, 1550–1556. doi: 10.7164/antibiotics.46.1550
Mason, C., Edwards, K., Carlson, R., Pignatello, J., Gleason, F., and Wood, J. (1982). Isolation of chlorine-containing antibiotic from the freshwater cyanobacterium Scytonema hofmanni. Science 215, 400–402. doi: 10.1126/science.6800032
Mazur-Marzec, H., Meriluoto, J., Plinski, M., and Szafranek, J. (2006). Characterization of nodularin variants in Nodularia spumigena from the Baltic Sea using liquid chromatography/mass spectrometry/mass spectrometry. Rapid Commun. Mass Spectrom. 20, 2023–2032. doi: 10.1002/rcm.2558
Meeks, J., Campbell, E., Summers, M., and Wong, F. (2002). Cellular differentiation in the cyanobacterium Nostoc punctiforme. Arch. Microbiol. 178, 395–403. doi: 10.1007/s00203-002-0476-5
Meeks, J. C., and Elhai, J. (2002). Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associateds ymbiotic growth states. Microbiol. Mol. Biol. Rev. 66, 94–121. doi: 10.1128/MMBR.66.1.94-121.2002
Mehdy, M. C. (1994). Active oxygen species in plant defense against pathogens. Plant Physiol. 105, 467–472.
Mikalsen, B., Boison, G., Skulberg, O. M., Fastner, J., Davies, W., Gabrielsen, T. M., et al. (2003). Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. J. Bacteriol. 185, 2774–2785. doi: 10.1128/JB.185.9.2774-2785.2003
Moore, R. E., Cheuk, C., and Patterson, G. M. L. (1984). Hapalindoles: new alkaloids from the blue-green alga Hapalosiphon fontinalis. J. Am. Chem. Soc. 106, 6456–6457. doi: 10.1021/ja00333a079
Mortier, K. A., Zhang, G. F., van Peteghem, C. H., and Lambert, W. E. (2004). Adduct formation in quantitative bioanalysis: effect of ionization conditions on paclitaxel. J. Am. Soc. Mass. Spectrom. 15, 585–592. doi: 10.1016/j.jasms.2003.12.013
Neefs, J. M., Van De Peer, Y., Hendriks, L., and De Wachter, R. (1990). Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 18, 2237–2317. doi: 10.1093/nar/18.suppl.2237
Nilsson, M., Rasmussen, U., and Bergman, B. (2005). Competition among symbiotic cyanobacterial Nostoc strains forming artificial associations with rice (Oryza sativa). FEMS Microbiol. Lett. 245, 139–144. doi: 10.1016/j.femsle.2005.03.010
Oksanen, I., Jokela, J., Fewer, D. P., Wahlsten, M., Rikkinen, J., and Sivonen, K. (2004). Discovery of rare and highly toxic microcystins from lichen-associated cyanobacterium Nostoc sp. strain IO-102-I. Appl. Environ. Microbiol. 70, 5756–5763. doi: 10.1128/AEM.70.10.5756-5763.2004
Papaefthimiou, D., Hrouzek, P., Mugnai, M. A., Lukesova, A., Turicchia, S., Rasmussen, U., et al. (2008a). Differential patterns of evolution and distribution of the symbiotic behaviour in nostocacean cyanobacteria. Int. J. Syst. Evol. Microbiol. 58, 553–564. doi: 10.1099/ijs.0.65312-0
Papaefthimiou, D., Van Hove, C., Lejeune, A., Rasmussen, U., and Wilmotte, A. (2008b). Diversity and host specificity of Azolla cyanobionts. J. Phycol. 44, 60–70. doi: 10.1111/j.1529-8817.2007.00448.x
Pluotno, A., and Carmeli, S. (2005). Banyasin A and banyasides A and B, three novel modified peptides from a water bloom of the cyanobacterium Nostoc sp. Tetrahedron 61, 575–583. doi: 10.1016/j.tet.2004.11.016
Rantala, A., Fewer, D. P., Hisbergues, M., Rouhiainen, L., Vaitomaa, J., Borner, T., et al. (2004). Phylogenetic evidence for the early evolution of microcystin synthesis. Proc. Natl. Acad. Sci. U.S.A. 101, 568–573. doi: 10.1073/pnas.0304489101
Rasmussen, U., and Svenning, M. (2001). Characterization by genotypic methods of symbiotic Nostoc strains isolated from five species of Gunnera. Arch. Microbiol. 176, 204–210. doi: 10.1007/s002030100313
Rasmussen, U., and Svenning, M. M. (1998). Fingerprinting of cyanobacteria based on PCR with primers derived from short and long tandemly repeated repetitive sequences. Appl. Environ. Microbiol. 64, 265–272.
Ray, S., and Bagchi, S. N. (2001). Nutrients and pH regulate algicide accumulation in cultures of the cyanobacterium Oscillatoria laetevirens. New Phytol. 149, 455–460. doi: 10.1046/j.1469-8137.2001.00061.x
Reinhold, B. B., Hauer, C. R., Plummer, T. H., and Reinhold, V. N. (1995). Detailed structural analysis of a novel, specific O-linked glycan from the prokaryote Flavobacterium meningosepticum. J. Biol. Chem. 270, 13197–13203. doi: 10.1074/jbc.270.22.13197
Rezanka, T., and Dembitsky, V. M. (2006). Metabolites produced by cyanobacteria belonging to several species of the family Nostocaceae. Folia Microbiol. 51, 159–182. doi: 10.1007/BF02932119
Rodgers, G. A., and Stewart, W. D. P. (1977). The cyanophyte-hepatic symbiosis. I. Morphology and physiology. New Phytol. 78, 441–458.
Rouhiainen, L., Jokela, J., Fewer, D. P., Urmann, M., and Sivonen, K. (2010). Two alternative starter modules for the non-ribosomal biosynthesis of specific anabaenopeptin variants in Anabaena (Cyanobacteria). Chem. Biol. 17, 265–273. doi: 10.1016/j.chembiol.2010.01.017
Schindler, C. S., Bertschi, L., and Carreira, E. M. (2010). Total synthesis of nominal banyaside B: structural revision of the glycosylation site. Angew. Chem. Int. Ed. Engl. 49, 9229–9232. doi: 10.1002/anie.201004047
Schloss, P. D., and Handelsman, J. (2005). Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71, 1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005
Singh, R. K., Tiwari, S. P., Rai, A. K., and Mohapatra, T. M. (2011). Cyanobacteria: an emerging source for drug discovery. J. Antibiot. (Tokyo) 64, 401–412. doi: 10.1038/ja.2011.21
Sivonen, K., Carmichael, W. W., Namikoshi, M., Rinehart, K. L., Dahlem, A. M., and Niemela, S. I. (1990). Isolation and characterization of hepatotoxic microcystin homologs from the filamentous freshwater cyanobacterium Nostoc sp. strain 152. Appl. Environ. Microbiol. 56, 2650–2657.
Sprent, J. I., and Meeks, J. C. (2013). Cyanobacterial nitrogen fixation in association with feather mosses: moss as boss? New Phytol. 200, 5–6. doi: 10.1111/nph.12449
Stanier, R. Y., Kunisawa, R., Mandel, M., and Cohen-Bazire, G. (1971). Purification and properties of unicellular bluegreen algae (Order Chroococcales). Bacteriol. Rev. 35, 171–205.
Svenning, M. M., Eriksson, T., and Rasmussen, U. (2005). Phylogeny of symbiotic cyanobacteria within the genus Nostoc based on 16S rDNA sequence analyses. Arch. Microbiol. 183, 19–26. doi: 10.1007/s00203-004-0740-y
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Tan, L. T. (2013). Pharmaceutical agents from filamentous marine cyanobacteria. Drug Discov. Today 18, 863–871. doi: 10.1016/j.drudis.2013.05.010
Thanh Doan, N., Rickards, R. W., Rothschild, J. M., and Smith, G. D. (2000). Allelopathic actions of the alkaloid 12-epi-hapalindole E isonitrile and calothrixin A from cyanobacteria of the genera Fischerella and Calothrix. J. Appl. Phycol. 12, 409–416. doi: 10.1023/A:1008170007044
Todorova, A. K., Juettner, F., Linden, A., Pluess, T., and von Philipsborn, W. (1995). Nostocyclamide: a new macrocyclic, thiazole-containing allelochemical from Nostoc sp. 31 (cyanobacteria). J. Org. Chem. 60, 7891–7895. doi: 10.1021/jo00129a032
Tooming-Klunderud, A., Mikalsen, B., Kristensen, T., and Jakobsen, K. S. (2008). The mosaic structure of the mcyABC operon in Microcystis. Microbiology 154, 1886–1899. doi: 10.1099/mic.0.2007/015875-0
Vardi, A., Schatz, D., Beeri, K., Motro, U., Sukenik, A., Levine, A., et al. (2002). Dinoflagellate-cyanobacterium communication may determine the composition of phytoplankton assemblage in a mesotrophic lake. Curr. Biol. 12, 1767–1772. doi: 10.1016/S0960-9822(02)01217-4
Walsh, C. T., Chen, H. W., Keating, T. A., Hubbard, B. K., Losey, H. C., Luo, L. S., et al. (2001). Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr. Opin. Chem. Biol. 5, 525–534. doi: 10.1016/S1367-5931(00)00235-0
Wang, H., Fewer, D. P., and Sivonen, K. (2011). Genome mining demonstrates the widespread occurrence of gene clusters encoding bacteriocins in cyanobacteria. PLoS ONE 6:e22384. doi: 10.1371/journal.pone.0022384
Waters, C. M., and Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Welker, M., and von Dohren, H. (2006). Cyanobacterial peptides - Nature’s own combinatorial biosynthesis. FEMS Microbiol. Rev. 30, 530–563. doi: 10.1111/j.1574-6976.2006.00022.x
West, N. J., and Adams, D. G. (1997). Phenotypic and genotypic comparison of symbiotic and free-living cyanobacteria from a single field site. Appl. Environ. Microbiol. 63, 4479–4484.
Wojtaszek, P. (1997). Oxidative burst: an early plant response to pathogen infection. Biochem. J. 322(Pt 3), 681–692. doi: 10.1042/bj3220681
Zheng, W., Bergman, B., Chen, B., Zheng, S., Guan, X., and Rasmussen, U. (2009). Cellular responses in the cyanobacterial symbiont during its vertical transfer between plant generations in the Azolla microphylla symbiosis. New Phytol. 181, 53–61. doi: 10.1111/j.1469-8137.2008.02644.x
Keywords: Cyanobacteria, secondary metabolites, symbiosis, Blasia, Nostoc, allelopathy
Citation: Liaimer A, Jensen JB and Dittmann E (2016) A Genetic and Chemical Perspective on Symbiotic Recruitment of Cyanobacteria of the Genus Nostoc into the Host Plant Blasia pusilla L. Front. Microbiol. 7:1693. doi: 10.3389/fmicb.2016.01693
Received: 21 July 2016; Accepted: 10 October 2016;
Published: 01 November 2016.
Edited by:
Christine Beemelmanns, Hans Knöll Institute, GermanyReviewed by:
Radha Prasanna, Indian Agricultural Research Institute (ICAR), IndiaErnani Pinto, University of São Paulo, Brazil
Josselin Noirel, Conservatoire National des Arts et Métiers, France
Copyright © 2016 Liaimer, Jensen and Dittmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anton Liaimer, anton.liaimer@uit.no
 Anton Liaimer
Anton Liaimer John B. Jensen
John B. Jensen Elke Dittmann
Elke Dittmann