Developing rapid DNA analysis technology

Jim Schneider, professor of chemical engineering at Carnegie Mellon University, recently received a 3-year, $295,000 grant from the National Science Foundation (NSF) to develop new strategies to rapidly separate and analyze long (kilobase) DNA strands, using surfactants rather than polymeric gels. These faster analysis techniques can impact a wide range of genomic DNA analyses, such as genome mapping, where rapid processing of kilobase-length DNA is required. While conventional pulsed-field electrophoresis methods take days to separate kilobase-length DNA, Schneider's new methods will take less than five minutes and use inexpensive, reusable materials.
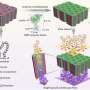
The key to Schneider's analysis method is the use of a soap-like nonionic surfactant material. The surfactant's slick, oily nature makes it a perfect alternative to the viscous polymeric gels that are currently a staple in electrophoretic DNA analysis. In electrophoresis, DNA is pulled through a buffer solution or gel with the help of an imposed electric field. This process separates the DNA, revealing a unique pattern of bands that can be used to characterize and identify DNA's genetic code.
"When DNA moves through the commonly used gel matrix, the DNA is forced to squeeze through tiny pores in the material. While this is required to separate the DNA, the high friction slows the process down," Schneider explains. "The gels formed by surfactants don't present permanent obstacles because of their dynamic, self-assembled structure. Instead, its [surfactants'] pores are constantly breaking apart and reforming, resulting in much lower friction and much faster runtimes."
Schneider is currently working to commercialize this technology, specifically for applications in forensic identification that involves genetic analysis. For example, Schneider hopes that his analysis methods can be implemented in medical diagnostics and crime labs.
"Investigators can use their existing equipment with our surfactant-based methods to perform DNA analysis ten to 100 times faster," says Schneider. "Unfortunately, investigators are often dealing with large backlogs of cases that require electrophoretic DNA analysis of field samples. We hope that this technology can really reduce these backlogs at a minimal expense."
Schneider has spent much of his career developing novel biomolecular sensing and separation methods that leverage principles of surfactant self-assembly to achieve fast runtimes and high sensitivity. Chemical engineering Ph.D. students Randall Gamble and Lingxiao Yan and M.S. student Ruohui Zheng are working with Schneider on this research.
For more information on Schneider's research, please read his previously published article in Analytical Chemistry, "A 502-base free-solution DNA sequencing method using end-attached micelles," and his article in Electrophoresis, "Optimization of ELFSE DNA sequencing with EOF counterflow and microfluidics."
Journal information: Analytical Chemistry